3.12 Multiple Populations
So far, we have only looked at simulations that involve one population. Adding multiple populations that interact with each other requires only a few additions to our code.
SLiM has a great way of visualizing your populations, accessed through the following menu:
3.12.1 Adding a new population
In past simulations, we’ve added a single population using the command sim.addSubpop();. To add multiple subpopulations, simply use the command multiple times, creating multiple populations with different names.
1 early() {
sim.addSubpop("p1", 500);
sim.addSubpop("p2", 3000);
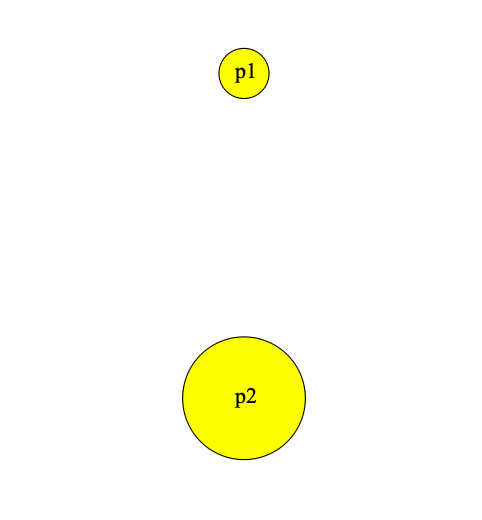
}Looking at our population viewer, we can see that there are now two populations displayed. The size of the circles in the viewer is proportionate to the size of the subpopulations that they represent.
3.12.2 Migration between populations
To set migration in SLiM, we can use <destination population>.setMigrationRates(<source population>, <rate>). An example of this function is: p1.setMigrationRates(p2, 0.2); The first population here, p1, is the population into which we migrate. The population in the parentheses is the population from which we migrate. The final number is the migration rate.
Adding migration to our simulation:
1 early() {
sim.addSubpop("p1", 500);
sim.addSubpop("p2", 3000);
p1.setMigrationRates(p2, 0.2);
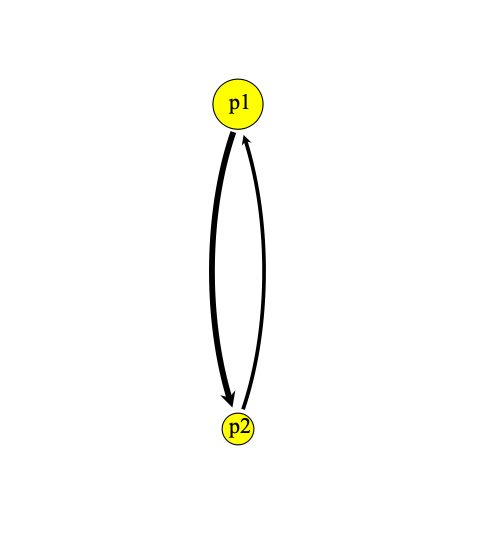
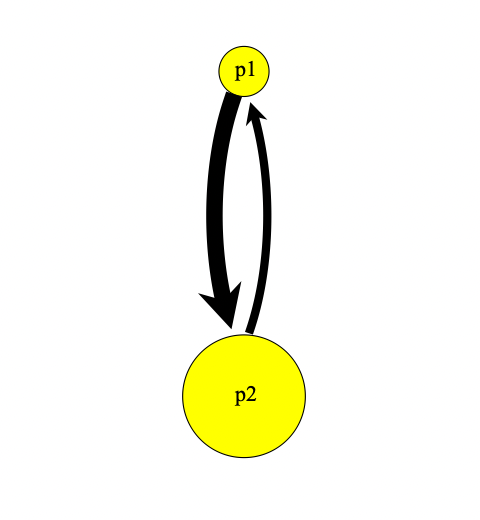
}Looking at our population visualization, we can see migration represented by an arrow:
If we add reverse migration, we can see that the size of the arrow is proportionate to the migration rate
1 early() {
sim.addSubpop("p1", 500);
sim.addSubpop("p2", 3000);
p1.setMigrationRates(p2, 0.2);
p2.setMigrationRates(p1, 0.8);
}
Note that we can use multiple calls of setMigrationRates() to change the migration rate throughout our simulation, as such:
1 early() {
sim.addSubpop("p1", 500);
sim.addSubpop("p2", 3000);
p1.setMigrationRates(p2, 0.2);
}
15 early() {
p1.setMigrationRates(p2, 0.4);
}3.12.3 Splitting a population into two
Real biological populations have common origins, rather than originating independently. We can incorporate this into our SLiM simulations by creating a new population as an offshoot of an existing population. This is accomplished with the function sim.addSubpopSplit(<new pop name>, <new pop size>, <source pop>)
For example, we can initialize a population with 500 individuals as such:
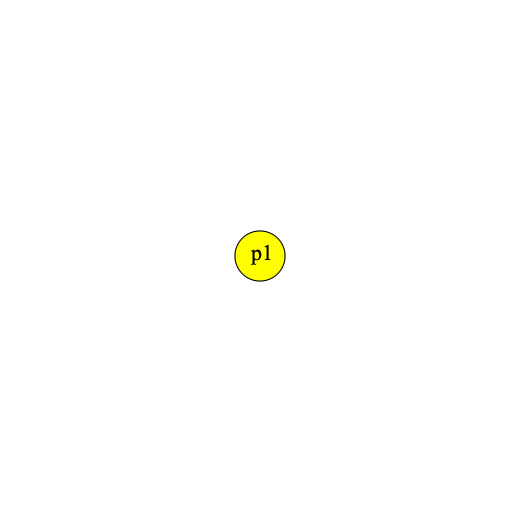
1 early() {
sim.addSubpop("p1", 500);
}Our population viewer looks like this:
We can add a population split as such:
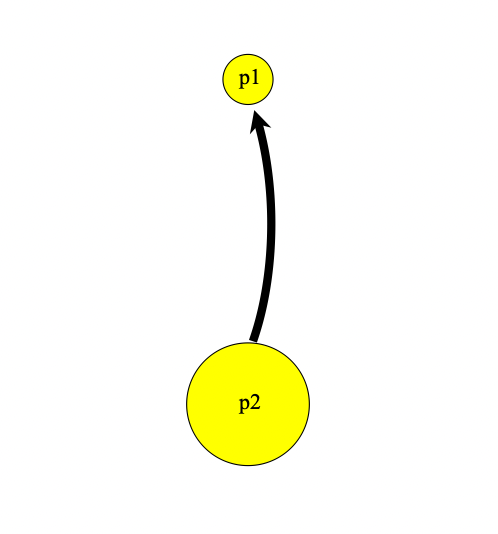
300 early() {
sim.addSubpopSplit("p2", 200, p1);
p2.setMigrationRates(p1, 0.1);
p1.setMigrationRates(p2, 0.04);
}Here we create a population p2, derived from population p1 with 200 individuals. We subsequently set weak migration rates between the populations. Our population viewer now looks like this: